Research
The aim of our research is to uncover how respiratory viral populations are shaped by adaptive immune responses and to use this information to predict and manage their coevolution through vaccination.
Evolution of the antibody response
A host's primary means of protection to pathogens such as influenza and SARS-CoV-2 is through antibodies, which themselves evolve rapidly by natural selection as B cell receptors. Understanding the dynamics of antibody repertoires is thus key to understanding much pathogen evolution and variation in susceptibility between people. We are investigating the evolutionary dynamics of primary and secondary responses, in part to measure the strength of competition between pre-existing and new responses (phenomena known as "original antigenic sin" or "immune imprinting").
Relevant papers:- Vieira, M.C., Palm, A.E., Stamper, C.T., Tepora, M.E., Nguyen, K.D., Pham, T.D., Boyd, S.D., Wilson, P.C. and S. Cobey. 2023. Germline-encoded specificities and the predictability of the B cell response. PLOS Pathogens 19(8): e1011603
- Vieira, M., Zinder, D. and S. Cobey. 2018. Selection and neutral mutations drive pervasive mutability losses in long-lived B cell lineages. Molecular Biology and Evolution 35(5):1135-1146.
- Cobey, S., Wilson, P. and F. Matsen. 2015. The evolution within us. Philosophical Transactions of the Royal Society B 370(1676): 20140235.
- Childs, L., Baskerville, E. and S. Cobey. 2015. Tradeoffs in antibody repertoires to complex antigens. Philosophical Transactions of the Royal Society B. 370(1676): 20140245.
The dynamics of protection
Understanding the dynamics of protection is critical to managing viral populations and designing effective vaccines. We showed that the first infecting, or “imprinting,” subtype of influenza A cuts the risk of future clinical infection with related subtypes roughly in half for life, and the age distributions of infections with influenza B reflect similar imprinting effects. We used latent-state models to estimate that adults and children are protected for several years after infection, but this protection seems to derive from different sources, with antibody titers to the top of the hemagglutinin a better correlate of protection in children compared to adults. This might reflect immune memory boosting less-neutralizing responses and biasing toward conserved epitopes. With diverse longitudinal cohorts, we are estimating the durability of protection and its relationship to viral antigenic evolution and infection history.
Relevant papers:- Vieira, M.C., Donato, C.M., Arevalo, P., Rimmelzwaan, G.F., Wood, T., Lopez, L., Huang, Q.S., Dhanasekaran, V., Koelle, K. and S. Cobey. 2021. Lineage-specific protection and immune imprinting shape the age distributions of influenza B cases. Nature Communications 12: 4313
- Arevalo, P., McLean, H.Q., Belongia, E.A. and S. Cobey. 2020. Earliest infections predict the age distribution of influenza A cases. eLife 2020;9:e50060
- Ranjeva, S., Subramanian, R., Fang, Vicky J., Leung, Gabriel M., Ip, Dennis K. M., Perera, Ranawaka A. P. M., Peiris, J. S. Malik, Cowling, Benjamin J., and S. Cobey. 2019. Age-specific differences in the dynamics of protective immunity to influenza. Nature Communications 10:1660
Immunogenicity and effectiveness of the seasonal influenza vaccine
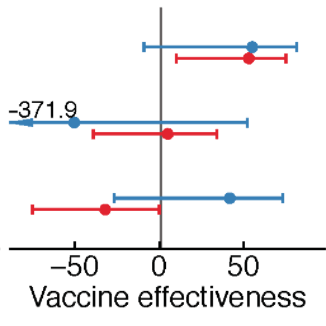
The low effectiveness of the seasonal influenza vaccine is often attributed to poor strain selection due to influenza's fast evolution. In practice, influenza vaccines often do not induce strong immune responses to the strains they do contain, and effectiveness appears low even in years with a good match. Using observational cohorts and a randomized clinical trial, we are investigating how past infections and vaccinations influence the immune response to vaccination. We are also investigating how estimates of vaccine effectiveness are influenced by uncertainty in the amount of immunity in unvaccinated populations, and we are developing methods to separate the protection conferred by vaccination from infection.
Relevant papers:- S. Cobey. 2024. Vaccination against rapidly evolving pathogens and the entanglements of memory. Nature Immunology 25(11): 2015-2023
- Cowling, B.J., Wong, S., Santos, J.J.S., Touyon, L., Ort, J..... Hensley, S.E. and S. Cobey. 2024. Preliminary findings from the Dynamics of the Immune Responses to Repeat Influenza Vaccination Exposures (DRIVE I) Study: a Randomized Controlled Trial. Clinical Infectious Diseases 79(4): 901-909
- Bi, Q., Dickerman, B.A., McLean, H.Q., Martin, E.T., Gaglani, M., Wernli, K.J., Goundappa, B., Flannery, B., Lipsitch, M. and S. Cobey and US Flu Vaccine Effectiveness Network Investigators. 2024. Reduced effectiveness of repeat influenza vaccination: distinguishing among within-season waning, recent clinical infection, and subclinical infection. The Journal of Infectious Diseases jiae220
- Cobey, S., Gouma, S., Parkhouse, K., Chambers, B.S., Ertl, H.C., Schmader, K. E., Halpin, R.A., Lin, X., Stockwell, T. B., Das, S. R., Landon, E., Tesic, V., Youngster, I., Pinsky, B., Wentworth, D.E., Hensley, S.E. and Y.H. Grad. 2018. Poor immunogenicity, not vaccine strain egg adaptation, may explain the low H3N2 influenza vaccine effectiveness in 2012-2013. Clinical Infectious Diseases 67(3):327-333
- Zost, S.J., Parkhouse, K., Gumina, M.E., Kim, K., Perez, S.D., Wilson, P.C., Treanor, J.J., Sant, A.J., Cobey, S. and S. Hensley. 2017. Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains. PNAS 114(47):12578-12583.
Ecology and evolution of influenza and other multistrain pathogens
Many pathogens comprise multiple strains that appear somewhat distinct to the immune system. Protective immunity can be a strong evolutionary pressure, as in influenza, and it can also stabilize the dynamics of competing strains and promote coexistence. We are investigating how immune pressure interacts with strains' transmission dynamics and evolution to shape pathogen diversity. Understanding the interplay of these forces is useful for the anticipating the evolutionary impacts of vaccination strategies, which can alter the strength and type of immune selection. It can also help us understand variation in epidemic sizes in space and time.
Relevant papers:- McGough, L. and S. Cobey. 2024. A speed limit on serial strain replacement from original antigenic sin. PNAS 121(25): e2400202121
- Kim, K., Gouma, S., Vieira, M.C., Weirick, M., Hensley, S.E. and S. Cobey. Measures of population immunity can predict the dominant clade of influenza A (H3N2) in the 2017-2018 season and reveal age-associated differences in susceptibility and antibody-binding specificity. 2024. Influenza and Other Respiratory Viruses 18(11): e70033
- Wen, F., Malani, A. and S. Cobey. 2022. The potential beneficial effects of vaccination on antigenically evolving pathogens. American Naturalist 199(2): 223-227
- Ranjeva, S., Baskerville, E., Dukic, V., Villa, L., Lazcano-Ponce, E., Giuliano, A., Dwyer, G. and S. Cobey. 2017. Recurring infection with ecologically distinct human papillomavirus (HPV) types explains high prevalence and diversity. PNAS 114(51):13573-13578.
- Wen, F., Bedford, T. and S. Cobey. 2016. Explaining the geographic origins of seasonal influenza A (H3N2). Proceedings of the Royal Society B. 283(1838).
Assistance with the early COVID-19 response
In 2020 and 2021, we developed statistical tools and models to provide situational awareness and inform interventions to SARS-CoV-2. Some of this work involved Illinois-specific analyses, which we regularly communicated to state leaders. We also published general papers on Rt estimation and vaccination strategies. Our work led to consultations with federal government agencies, civic organizations, and the WHO, but most of it remains unpublished.
- Cobey, S. 2020. Modeling infectious disease dynamics. Science 368(6492):713-714.
- Gostic, K.M., McGough, L., Baskerville, E., Abbott, S., .... S. Cobey 2020. Practical considerations for measuring the effective reproductive number, Rt. PLOS Computational Biology 16(12):e1008409
- Cobey, S., et al. 2021. Concerns about SARS-CoV-2 evolution should not hold back efforts to expand vaccination. Nature Reviews Immunology 21(5) 330-335.
- Bloom, J., et al. 2021. Investigate the origins of COVID-19. Science 372(6543): 694.
- Bubar, K.M., Reinholt, K., Kissler, S.M., Lipsitch, M., Cobey, S., Grad, Y.H. and D. B. Larremore. 2021. Model-informed COVID-19 vaccine prioritization by age and serostatus. Science 371(6532):916-921
- Holden, T., et al. 2021. Geographic and demographic heterogeneity of SARS-CoV-2 diagnostic testing in Illinois, USA, March to December 2020. BMC Public Health 21(1): 1-13.
- Cowling, B.J., Lim, W.W. and S. Cobey. 2021. Fractionation of COVID-19 vaccine doses could extend limited supplies and reduce mortality. Nature Medicine 27: 1321-1323.